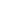Microscope Photography and Image Processing(ImageStudio)
a. Tools Introduction
ImageStudio is an offline image processing software for STOmics, mainly involving modules for image quality assessment and manual adjustment of images (image stitching, tissue segmentation, and cell segmentation). Image quality assessment primarily scores images captured by microscopes based on two evaluation metrics: image clarity and track line, used to determine if they meet the requirements for downstream data analysis. The manual adjustment module for images addresses scenarios where automated stitching or segmentation algorithms are insufficient, allowing manual tools to process the images.
b. Function Introduction
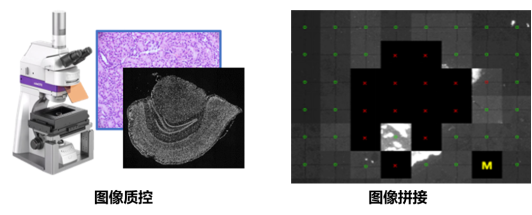
1. Image Quality Control
The image quality control module primarily evaluates whether the microscope images in spatiotemporal omics projects meet the requirements for subsequent data analysis. The module mainly includes three major functions: "ssDNA/DAPI" and DAPI&mIF image QC, uploading of images in TIFF/PNG formats, and configuration data uploading methods.
2. Image Stitching
The image stitching module is based on the chip's own track lines and is used to manually adjust the stitching effect of microscope thumbnails. The input files should be microscope thumbnail files with passing image QC results.
3. Image Calibration
The image calibration module is used to manually adjust the alignment of proteins and DAPI in mIF, primarily in the horizontal and vertical directions. The input files should be data obtained after DAPI&mIF image QC.
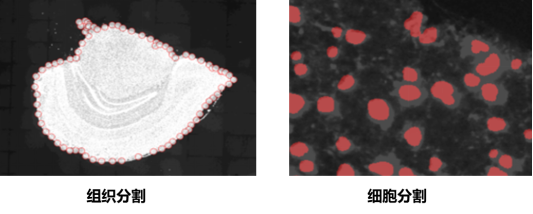
4. Tissue Segmentation
The tissue segmentation module is used to manually adjust the segmented tissue mask.