The research object of Transcriptome sequencing is the sum of all RNA that can be transcribed by a specific cell under a certain functional state. Current sequencing technology mainly targets mRNA. Transcriptome sequencing technology can obtain the issue of changes in gene expression levels, discovery of rare transcripts, precise identification of alternative splicing sites, and gene fusions, provide the most comprehensive transcriptome information. Through sequencing, we can obtain almost all transcript sequence information of a specific tissue or organ of a specie under a certain state. which is widely used in species genetic development, improvement, and molecular breeding research.

Library construction typically requires a minimum of 200 ng.
Different library construction strategy can be customized for special samples.
Extensive experience in library construction for pg-level trace samples and difficult-to-extract samples.

Rolling circle amplification constructs DNB sequencing library, PCR-free resequencing detects InDels more accurately. No worries about index hopping. Low duplication rate without manual intervention.

DNBSEQ-T7 and DNBSEQ-T20 have ultra-high throughput, short cycles, and high cost-effectiveness.
Applications

The Physiological Mechanisms of Growth and Development

Research on the Pathogenesis of Microbial Infection

Complex Diseases Research

Cancer Research and Biomarkers

Drug action Mechanism and Target Point
Immune Response, Stem Cell Research

Molecular Markers
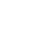
Species Comprehensive Transcriptome Atlas

Adaptive Mechanisms Research in Animals and Plants

Species Evolution Research
Technical procedure